Curate & link spatial data#
Show code cell content
!lamin init --storage ./test-spatial --schema bionty
💡 creating schemas: core==0.45.5 bionty==0.29.6
✅ saved: User(id='DzTjkKse', handle='testuser1', email='testuser1@lamin.ai', name='Test User1', updated_at=2023-08-17 14:13:09)
✅ saved: Storage(id='yLwg27uv', root='/home/runner/work/lamin-usecases/lamin-usecases/docs/test-spatial', type='local', updated_at=2023-08-17 14:13:09, created_by_id='DzTjkKse')
✅ loaded instance: testuser1/test-spatial
💡 did not register local instance on hub (if you want, call `lamin register`)
import lamindb as ln
import lnschema_bionty as lb
import matplotlib.pyplot as plt
import scanpy as sc
lb.settings.species = "human"
ln.settings.verbosity = 3
Show code cell output
✅ loaded instance: testuser1/test-spatial (lamindb 0.50.7)
✅ set species: Species(id='uHJU', name='human', taxon_id=9606, scientific_name='homo_sapiens', updated_at=2023-08-17 14:13:12, bionty_source_id='BH1c', created_by_id='DzTjkKse')
ln.track()
💡 notebook imports: lamindb==0.50.7 lnschema_bionty==0.29.6 matplotlib==3.7.2 scanpy==1.9.3
✅ saved: Transform(id='daeFs3PkquDW-R', name='Curate & link spatial data', short_name='spatial', stem_id='daeFs3PkquDW', version='draft', type=notebook, updated_at=2023-08-17 14:13:13, created_by_id='DzTjkKse')
✅ saved: Run(id='FOpXFSKcomgkye0HBj9e', run_at=2023-08-17 14:13:13, transform_id='daeFs3PkquDW-R', created_by_id='DzTjkKse')
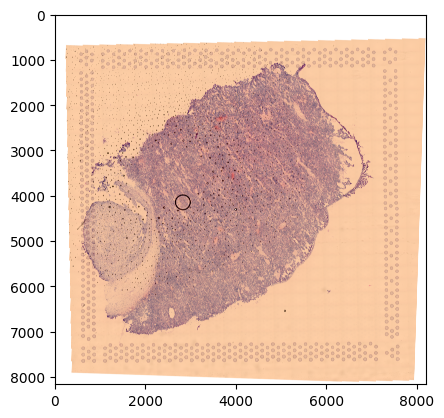
Here we have a spatial gene expression dataset measured using Visium from Suo22.
This dataset contains two parts:
a high-res image of a slice of fetal liver
a single cell expression dataset in .h5ad
img_path = ln.dev.datasets.file_tiff_suo22()
img = plt.imread(img_path)
plt.imshow(img)
plt.show()
adata = ln.dev.datasets.anndata_suo22_Visium10X()
# subset to the same image
adata = adata[adata.obs["img_id"] == "F121_LP1_4LIV"].copy()
adata
AnnData object with n_obs × n_vars = 3027 × 191
obs: 'in_tissue', 'array_row', 'array_col', 'sample', 'n_genes_by_counts', 'log1p_n_genes_by_counts', 'total_counts', 'log1p_total_counts', 'pct_counts_in_top_50_genes', 'pct_counts_in_top_100_genes', 'pct_counts_in_top_200_genes', 'pct_counts_in_top_500_genes', 'mt_frac', 'img_id', 'EXP_id', 'Organ', 'Fetal_id', 'SN', 'Visium_Area_id', 'Age_PCW', 'Digestion time', 'paths', 'sample_id', '_scvi_batch', '_scvi_labels', '_indices', 'total_cell_abundance'
var: 'feature_types', 'genome', 'SYMBOL', 'mt'
obsm: 'NMF', 'means_cell_abundance_w_sf', 'q05_cell_abundance_w_sf', 'q95_cell_abundance_w_sf', 'spatial', 'stds_cell_abundance_w_sf'
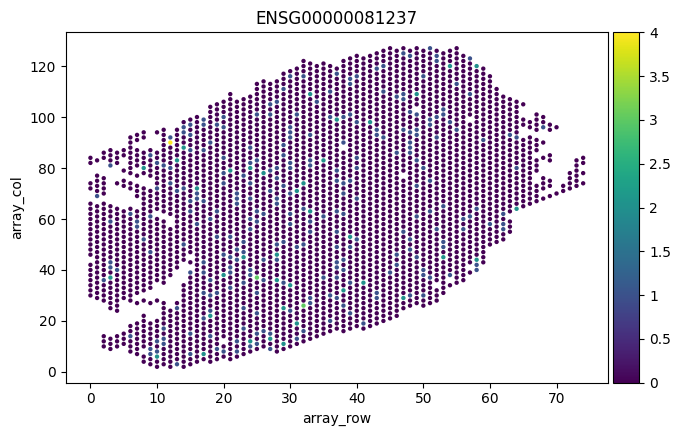
# plot where CD45+ leukocytes are in the slice
sc.pl.scatter(adata, "array_row", "array_col", color="ENSG00000081237")
Register the AnnData and image file as a dataset#
file_ad = ln.File.from_anndata(
adata,
description="Suo22 Visium10X image F121_LP1_4LIV",
var_ref=lb.Gene.ensembl_gene_id,
)
file_ad.save()
Show code cell output
💡 parsing feature names of X stored in slot 'var'
💡 using global setting species = human
✅ validated 191 Gene records from Bionty on ensembl_gene_id: ENSG00000002586, ENSG00000004468, ENSG00000004897, ENSG00000007312, ENSG00000008086, ENSG00000008128, ENSG00000010278, ENSG00000010610, ENSG00000012124, ENSG00000013725, ENSG00000019582, ENSG00000026508, ENSG00000039068, ENSG00000059758, ENSG00000062038, ENSG00000065883, ENSG00000066294, ENSG00000070831, ENSG00000071991, ENSG00000073754, ...
✅ linked: FeatureSet(id='PPJeyfVpv0w3oPkQciJS', n=191, type='float', registry='bionty.Gene', hash='vzXXh-48O_mReCwQfdJa', created_by_id='DzTjkKse')
💡 parsing feature names of slot 'obs'
❗ did not validate 27 Feature records for names: in_tissue, array_row, array_col, sample, n_genes_by_counts, log1p_n_genes_by_counts, total_counts, log1p_total_counts, pct_counts_in_top_50_genes, pct_counts_in_top_100_genes, pct_counts_in_top_200_genes, pct_counts_in_top_500_genes, mt_frac, img_id, EXP_id, Organ, Fetal_id, SN, Visium_Area_id, Age_PCW, ...
❗ ignoring non-validated features: in_tissue,array_row,array_col,sample,n_genes_by_counts,log1p_n_genes_by_counts,total_counts,log1p_total_counts,pct_counts_in_top_50_genes,pct_counts_in_top_100_genes,pct_counts_in_top_200_genes,pct_counts_in_top_500_genes,mt_frac,img_id,EXP_id,Organ,Fetal_id,SN,Visium_Area_id,Age_PCW,Digestion time,paths,sample_id,_scvi_batch,_scvi_labels,_indices,total_cell_abundance
❗ no validated features, skip creating feature set
✅ saved 1 feature set for slot: ['var']
✅ storing file 'wqo8zmAqIw4Ky2onlYdN' at '.lamindb/wqo8zmAqIw4Ky2onlYdN.h5ad'
file_img = ln.File(img_path, description="Suo22 image F121_LP1_4LIV")
file_img.save()
Show code cell output
✅ storing file '9xCpMQylCzk4gwFFMBP5' at '.lamindb/9xCpMQylCzk4gwFFMBP5.tiff'
dataset = ln.Dataset.from_files(files=[file_ad, file_img], name="Suo22")
dataset.save()
Show code cell content
# clean up test instance
!lamin delete --force test-spatial
!rm -r test-flow
💡 deleting instance testuser1/test-spatial
✅ deleted instance settings file: /home/runner/.lamin/instance--testuser1--test-spatial.env
✅ instance cache deleted
✅ deleted '.lndb' sqlite file
❗ consider manually deleting your stored data: /home/runner/work/lamin-usecases/lamin-usecases/docs/test-spatial
rm: cannot remove 'test-flow': No such file or directory